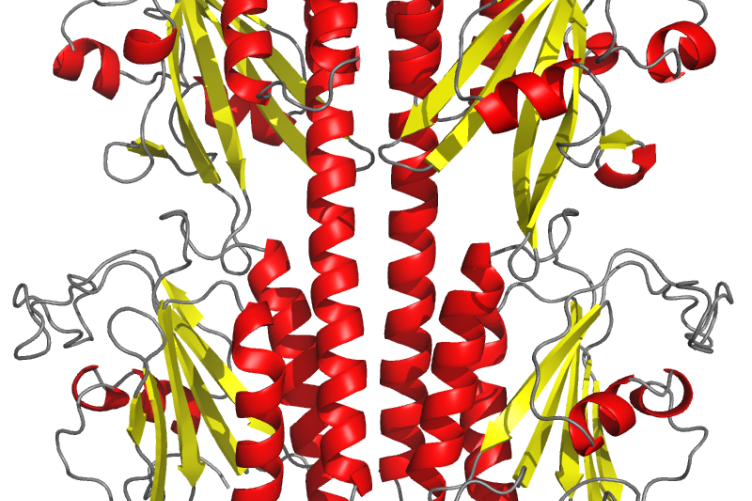
The PDE6 crystal structure used for the research. Photo credit: Richa Gupta
UNH researchers have advanced our understanding of the mutations that result in color blindness and certain retinal degenerative diseases that cause blindness. Their recently published study, led by UNH doctoral student Richa Gupta, takes an unprecedented look at the structure and function of phosphodiesterase-6 or PDE6 — a protein central to color vision in humans and often linked to retinal degenerative diseases — that’s found in cones, one of the eye’s two photoreceptors.
The outcome of their work, which included the development of the highest resolution crystallized image of PDE6 and the first atomic-level structural model of the enzyme in cones, provides the most detailed picture of how PDE6 is regulated during visual signaling in the retina and gives a molecular explanation for why certain PDE6 mutations cause vision problems, information that could lead to treatments that address the underlying cause of those conditions.
But the success of the study would not have been as far reaching without the combined interdisciplinary expertise that spanned three graduate programs and one undergraduate program at UNH.
At the outset of her study, Gupta worked with researchers Huanchen Wang, Wenjun Cui and Hengming Ke from the University of North Carolina at Chapel Hill to create an atomic level structure of the crystallized PDE6 protein. Independently, Gupta also created high resolution models of the protein when it is bound to regulatory molecules. While both are extremely valuable, they did not provide her with the data about PDE6 dynamics she was seeking.

Enter Chris Nordyke, a biochemistry doctoral student in the lab of Krisztina Varga, associate professor of molecular, cellular and biomedical sciences, and Yong Liu, a chemical engineering doctoral student in the lab of Harish Vashisth, associate professor of chemical engineering.
Nordyke specializes in nuclear magnetic resonance (NMR) spectroscopy, which is commonly used to understand the structure and motion of molecules and analyze complex chemical and biological systems. Along with assistance from undergraduate student Ryan Puterbaugh, he helped Gupta analyze the ways PDE6 binds to regulatory molecules and whether those reactions result in functional changes that might affect vision.
To further understand the molecular dynamics of protein motion, Gupta also worked with Liu, who created computer simulations of the atomic motions of PDE6 that illuminated how the enzyme’s structure changes when it binds to an intracellular messenger molecule. These molecular simulations start with the static structure of PDE6 and makes it come alive by letting the atoms move one femtosecond — one quadrillionth of a second — at a time, similar to creating drawings for an animated movie but at much shorter intervals.
It was the insight into the protein’s dynamics that produced the study’s key findings.
“By comparing the changes the enzyme undergoes when it binds to other molecules, Richa was able to identify functional attributes to regions that we didn’t know had a function, and some of these regions had PDE6 mutations,” says Rick Cote, director of the Center of Integrated Biomedical and Bioengineering Research and Gupta’s advisor, “So now we can better interpret the deleterious effects of these inherited mutations on the structure and function of PDE6, and we can better understand how these mutations cause visual dysfunction and disease.”
For Gupta, the research has given her a comprehensive understanding of how to study protein structure and dynamics, a knowledge that she and others can apply to any other work involving understanding the impacts of protein mutations.
“Getting that high-resolution structure, combined with our work developing different states of the protein, will definitely help other researchers who are studying PDE6,” she says. “But if there is any other disease where a mutation in the protein is responsible for the disease, we can definitely also apply this knowledge there.”
“It’s a wonderful demonstration of how graduate students from different projects, different laboratories, different expertise can talk to each other and come up with ideas that enrich their own projects and enrich their learning experience,” says Cote.
This work is supported by the National Institutes of Health with the goal of developing therapies that will restore and/or prolong normal vision in humans with inherited defects in the PDE6 protein.
-
Written By:
Sarah Schaier | College of Life Sciences and Agriculture