New research from UNH advances the understanding — and ultimately treatment — of a genetic mutation that causes vision disease and blindness.
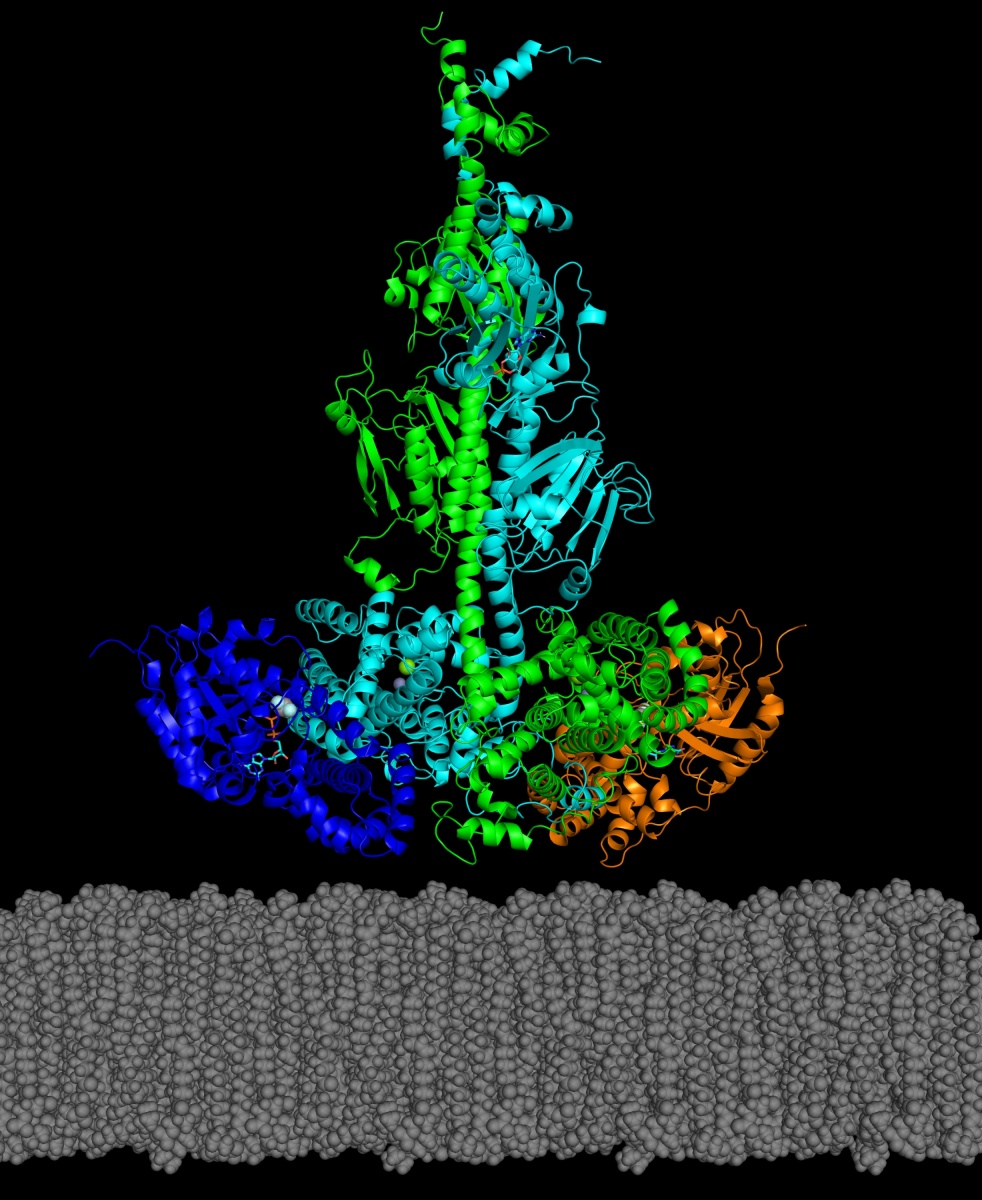
In a recent paper published in the Journal of Biological Chemistry, UNH researchers have reported the first structural model for a key enzyme, and its activating protein, that can play a role in some genetically inherited eye diseases like retinitis pigmentosa and night blindness.
“There has been substantial research on the biochemical pathway involving this enzyme, known as PDE6, but defining atomic-level models is important for locating PDE6 mutations in order to understand why they cause disease and how we can develop new therapeutic interventions to manage retinal diseases,” says Rick Cote, director of the Center of Integrated Biomedical and Bioengineering Research and principal investigator on the study.
Vision starts in the photoreceptor cells of the retina which contains rods, responsible for low light vision, and cones, which are active in brighter light and capable of color vision. When light is absorbed by the rods and cones, it triggers a pathway which activates the enzyme phosphodiesterase 6, or PDE6. This generates a nerve impulse to the brain that ultimately results in visual perception. Some genetically inherited eye diseases are caused by mutations to PDE6, or its activating protein, transducin, that can lead to disruptions of normal vision or even total blindness
"Defining atomic-level models is important for locating PDE6 mutations in order to understand why they cause disease and how we can develop new therapeutic interventions to manage retinal diseases.”
In the study, the researchers reported how they were able to use chemical cross-linking combined with mass spectrometric analysis to resolve the structure of PDE6 in its nonactivated and transducin-activated states. This approach permitted visualization of flexible regions of individual PDE6 catalytic and inhibitory subunits that were poorly resolved in previous work as well as the overall molecular architecture of the activated protein complex.
“Determining the structure of these visual signaling proteins has always been a challenge because of their complexity,” says Michael Irwin, doctoral student in biochemistry and lead author. “Having detailed structural information about how PDE6 is activated by transducin will help us understand the molecular causes of visual disorders and blinding diseases resulting from mutations in these proteins.”
Current medical treatment for such genetically inherited retina diseases may include gene therapy or drugs meant to inhibit the disease process. However, they are not always successful in restoring the balance of PDE6 and preventing blindness. Scientists believe that knowing the molecular structures of these visual signaling proteins and how they interact with each other can offer clues for the development of new drugs to both restore vision and prevent blindness.
This research was funded by the National Eye Institute, the National Institute of General Medical Sciences, the National Institute of Child Health and Human Development, the National Science Foundation and the UNH Research Office.
-
Written By:
Robbin Ray ’82 | UNH Marketing | robbin.ray@unh.edu | 603-862-4864